SERVICE
DNA-Seq
Metagenome(Shotgun Seq.)
Metagenome is a service that analyzes how many organisms (Bacteria, Fungi, etc.) exist in samples collected from various environments such as soil, air, water and human samples by applying NGS technology. Shotgun metagenome sequencing we provide is analyzing all genomic DNA in samples, not limited to specific regions (16S, 18S rRNA, ITS, etc.). by applying shotgun metagenome sequencing, It's possible to simultaneously identify and precisely profile various microorganisms such as bacteria, fungi and viruses.
Service Feature
|
Sample requirement |
>5µg gDNA, >50ng/ul gDNA, >4 DIN |
|
Library method |
NEB Ultra DNA Library kit |
|
NGS run format |
NovaSeq, PE150bp |
|
Data yield |
~10Gb/sample⁺ |
|
Turnaround time |
~4 weeks after Library QC |
|
Sample type |
gDNA⁺⁺ |
+ This is based on fecal samples, other sample types require separate consultation.
++ If you need gDNA preparation, you can proceed through consultation.
Progress
-
1
Library prep.
-
2
Library QC
-
3
NGS
-
4
Data Analysis
-
5
Report
Data Analysis
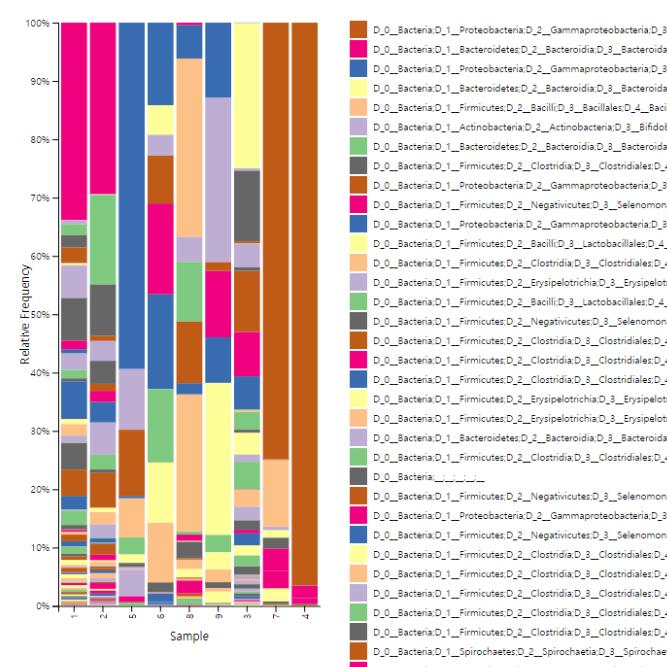
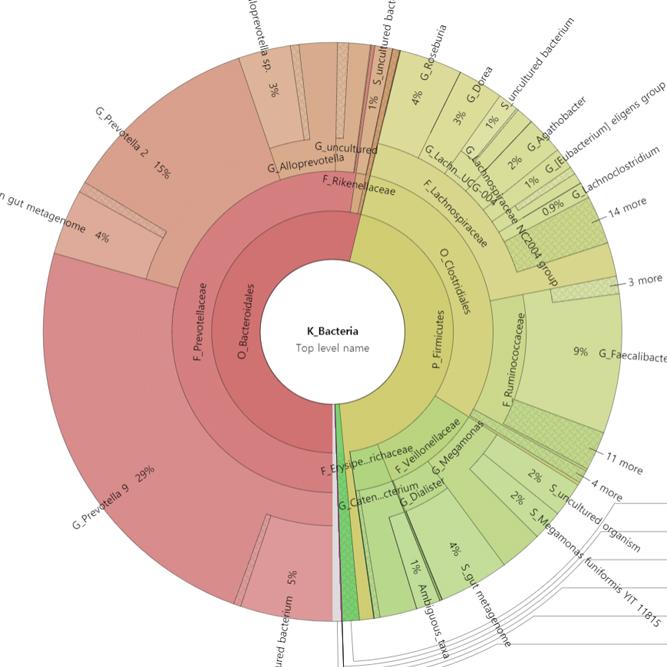
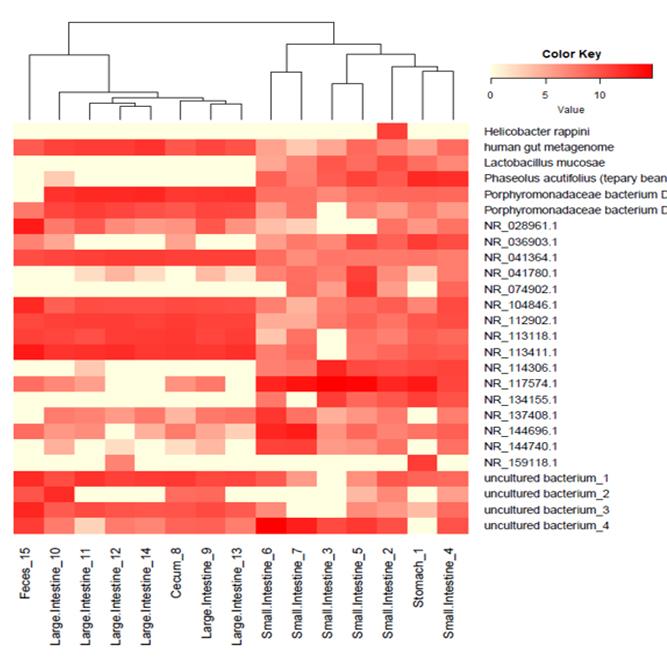
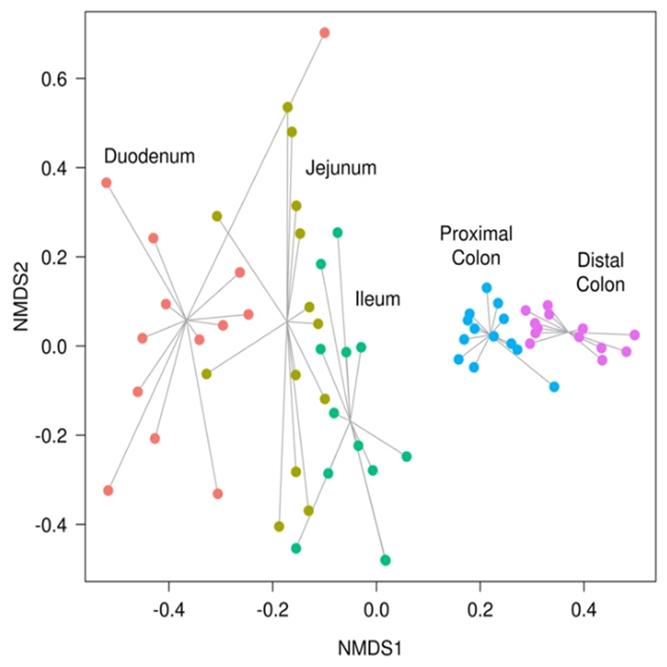
We support ExMEGA report, various analysis such as Assembly Result, Feature Table, Taxonomy(Bar & Pie Chart, Profile Data), Diversity(Alpha, Beta), Sample Clustering et cetera.