NGS(DNA/RNA-Seq)
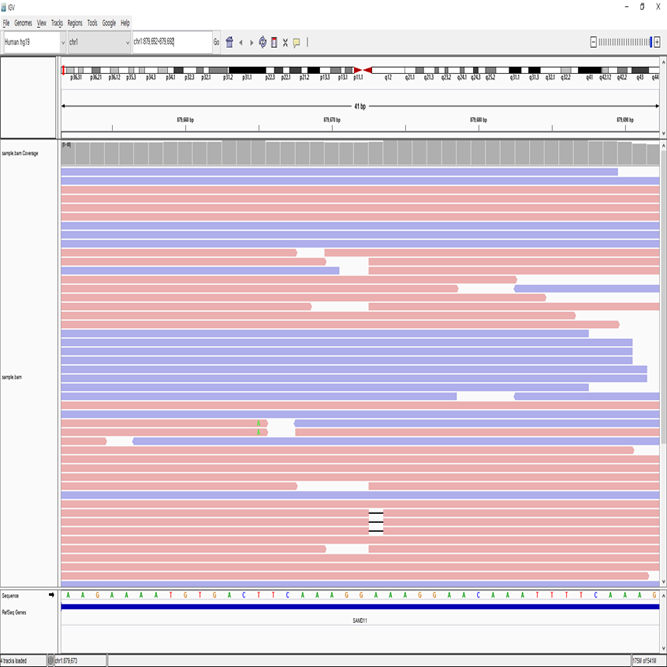
We provide whole genome sequencing service from gDNA samples using NGS technology. We report SNP, INDEL, CNV, Structural variation data and others
Service Info.
| Sample requirement | >2ug gDNA |
|---|---|
| Library method | Illumina TruSeq Nano DNA Library kit |
| NGS run format | NovaSeq 6000, PE150 |
| Data yield | ~90 Gb/sample |
| Turnaround time | ~7 weeks after DNA QC |
| Sample type | gDNA |
++ If you want to get DNA prep.service, you can send us your samples after discussing with us.
- DNA QC
- Library prep.
- NGS
- Data Analysis
- Report
Data Analysis
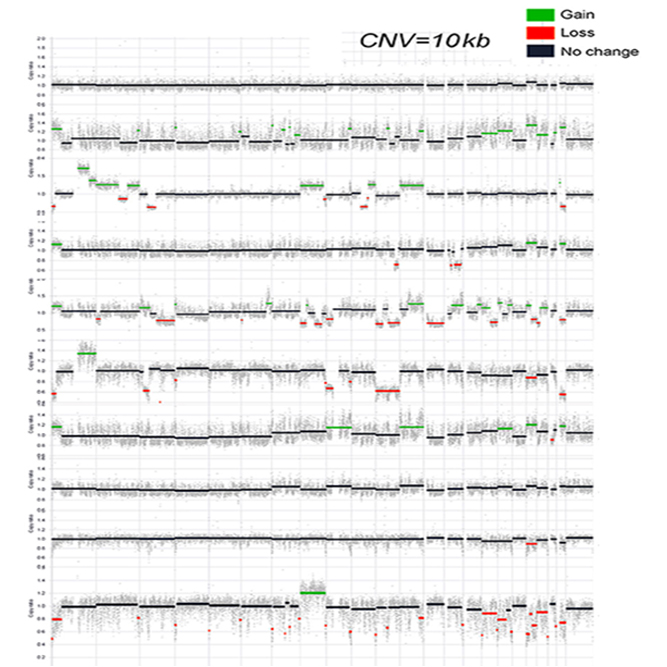
SNP(Single Nucleotide Polymorphism), INDEL (Insertion & Deletion), CNV(Copy Number Variation), Structural Variation analysis and Others
Whole Exome Sequencing is a method of analyzing genes in the Exom area by creating a library by selectively capturing only Exon areas. We report SNP, INDEL, CNV, Structural variation data and others
Service Info.
| Sample requirement | >2ug gDNA |
|---|---|
| Library method | Agilent SureSelect kit |
| NGS run format | NovaSeq 6000, PE100 |
| Data yield | ~6 Gb (100X depth)/sample |
| Turnaround time | ~6 weeks after DNA QC |
| Sample type | gDNA |
++ If you want to get DNA prep.service, you can send us your samples after discussing with us.
- DNA QC
- Library prep.
- NGS
- Data Analysis
- Report
Data Analysis
SNP(Single Nucleotide Polymorphism), INDEL (Insertion & Deletion), CNV(Copy Number Variation), Structural Variation analysis and Others
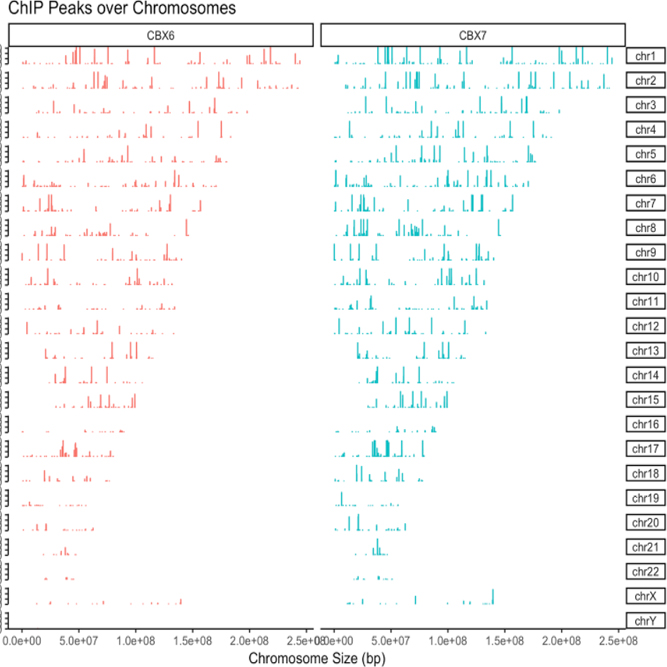
We provide considerably useful ChIP-Seq service for epigenetics study that analyzes the interaction of DNA and regulatory protein. With professional experiment know-hows and analysis technique developed over a long period of time, we provide data that meet the needs of a researcher.
Service Info.
| Sample requirement | >20ul IP-DNA |
|---|---|
| Library method | NEB Ultra DNA Library kit |
| NGS run format | Illumina NovaSeq6000, PE100bp |
| Data yield | ~4G/sample |
| Turnaround time | ~3 weeks after Library QC |
| Sample type | IP-DNA |
- Library Prep.
- Library QC
- NGS
- Data Analysis
- Report
Data Analysis
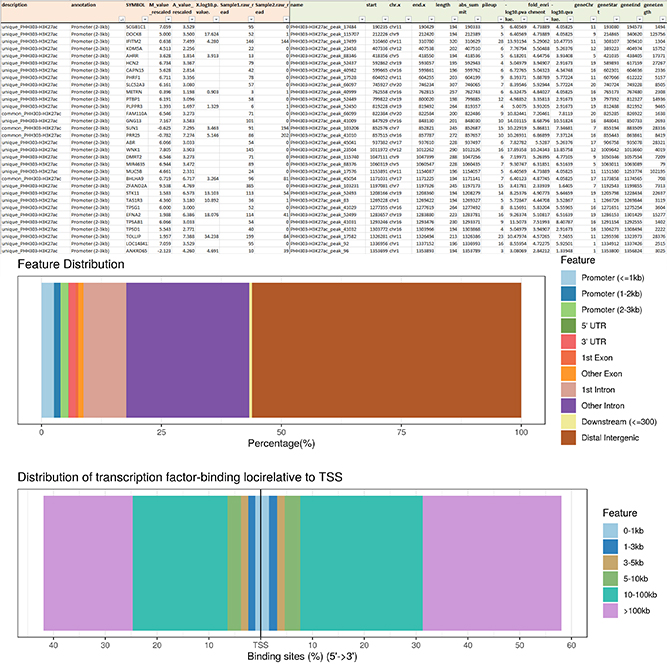
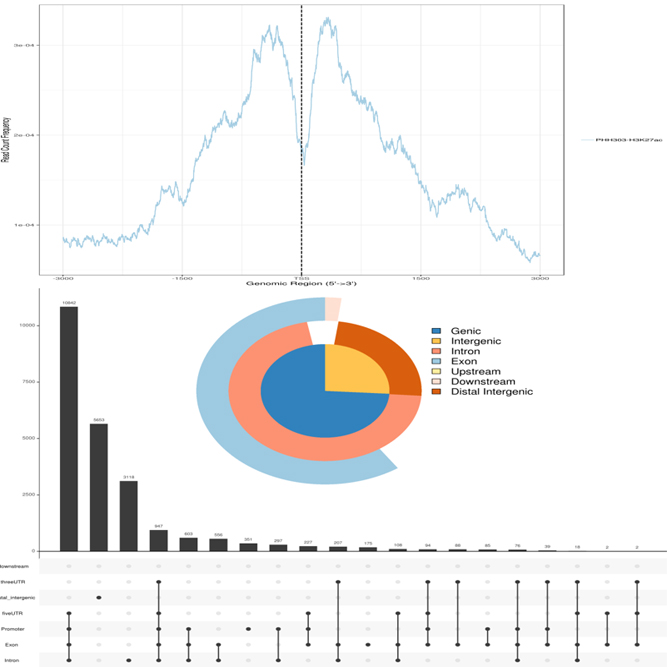
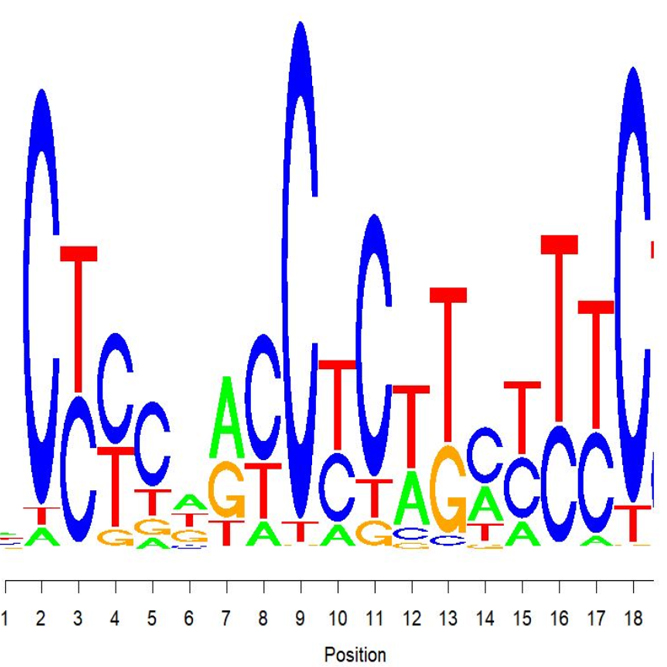
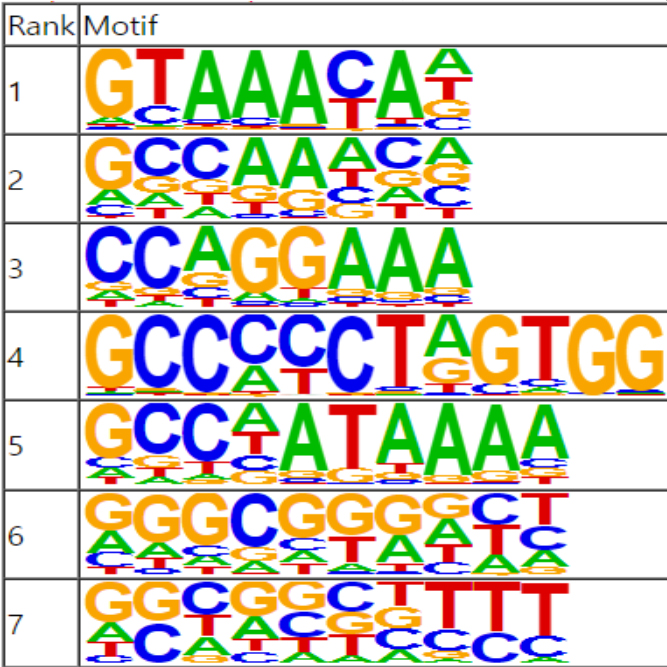
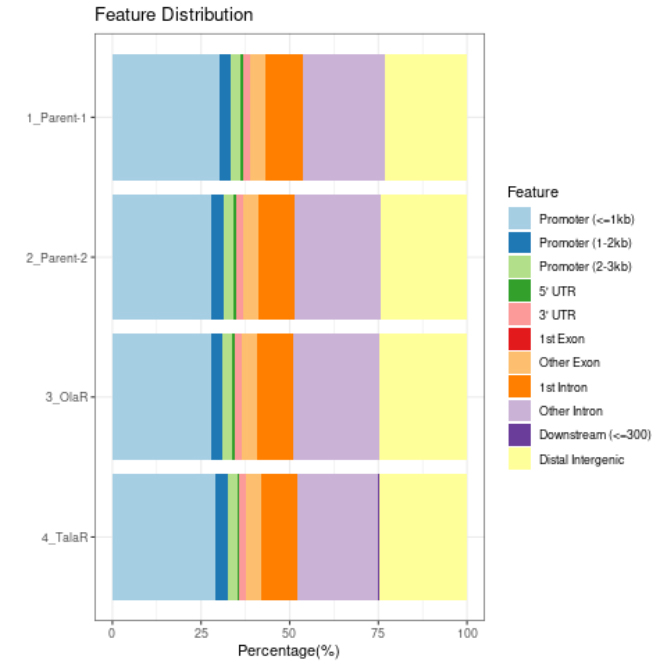
We support various analysis such as ExDEGA Report, Peak Annotation, Motif Discovery, Tags Comparison, IGV, BedGraph, Data Mining et cetera..
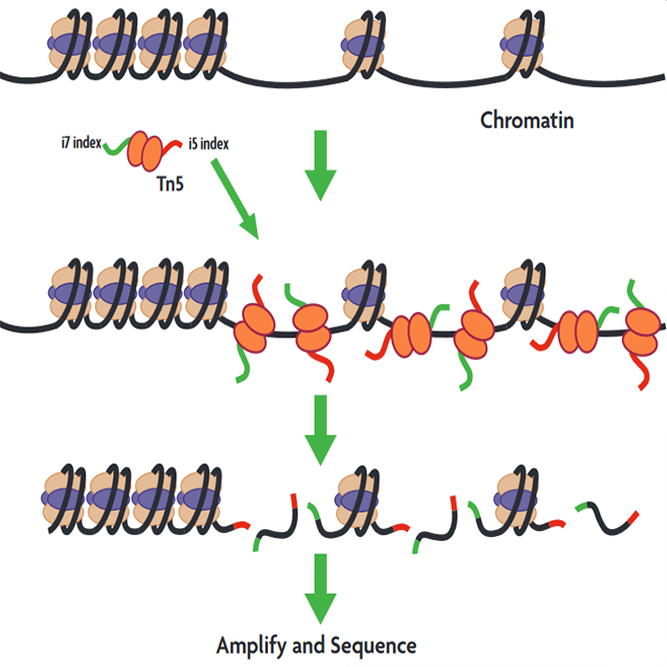
ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) is a technique used in molecular biology to assess genome-wide chromatin accessibility. ATAC-seq identifies accessible DNA regions by probing open chromatin with hyperactive mutant Tn5 Transposase that inserts sequencing adapters into open regions of the genome.
Service Info.
| Sample requirement | >~1x106 cells in 1ml media (Cell viability >80%) |
|---|---|
| Library method | ACTIVE MOTIF® ATAC-Seq Kit |
| NGS run format | NovaSeq 6000, PE100 |
| Data yield | ~6G/ sample |
| Turnaround time | ~5 weeks after Cell QC |
| Sample type | Cell |
- Cell QC
- Library prep.
- NGS
- Data Analysis
- Report
Data Analysis
We support various analysis such as ExPADA Report, Peak Annotation, Motif Discovery, Tags Comparison, IGV, BedGraph, Data Mining et cetera.
Provides metagenomic and microbiome analysis which identifies and quantifies microbial community (bacteria, fungi, etc) derived from various environments including soil, air, water, and human body by using 16s rRNA sequencing.
Service Info.
| Sample requirement | >0.1 ng/ul gDNA |
|---|---|
| Library method | 16S rRNA Amplicon Library for illumina |
| NGS run format | Illumina MiSeq, PE300bp(Paired End 300bp) |
| Data yield | ~50,000 reads/sample |
| Turnaround time | ~4 weeks after Library QC |
| Sample type | gDNA |
++ If you want to get DNA prep.service, you can send us your samples after discussing with us.
- DNA QC
- Library prep.
- NGS
- Data Analysis
- Report
Data Analysis
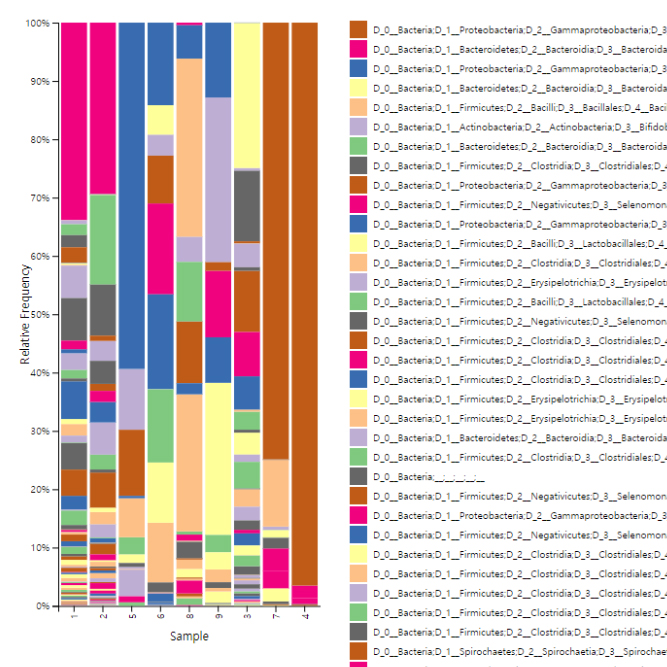
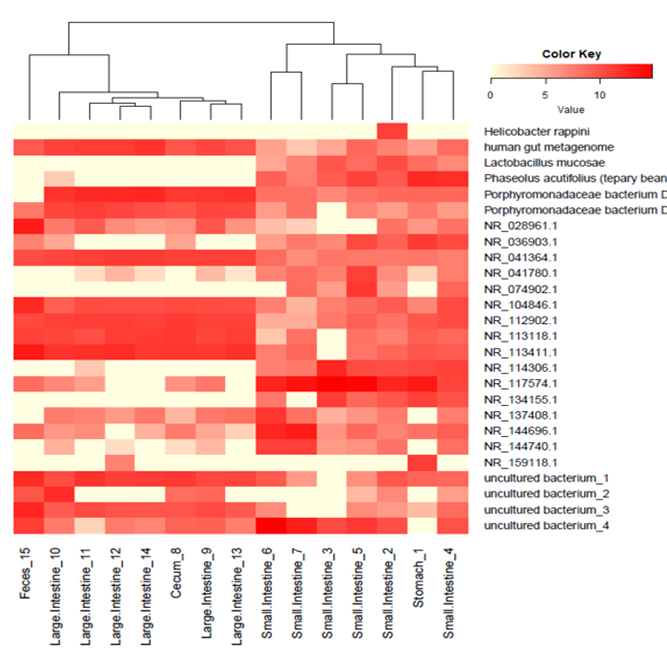
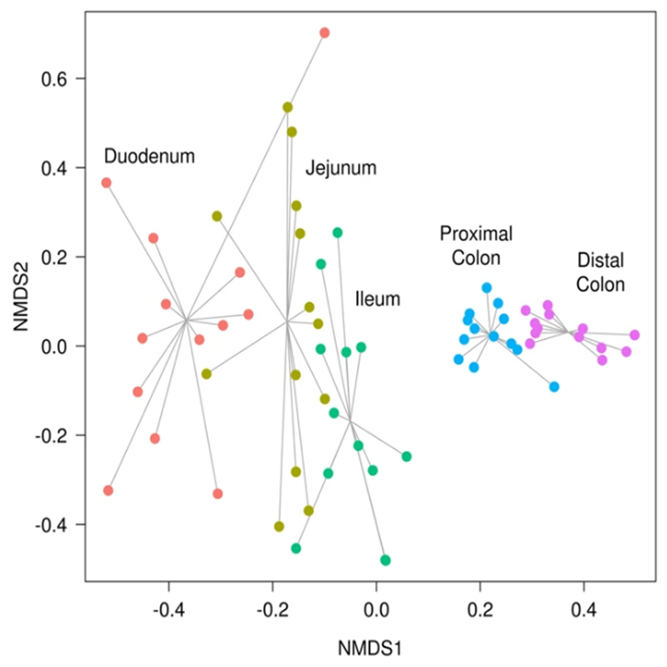
We support various analysis such as Assembly Result, Feature Table, Taxonomy(Bar & Pie Chart, Profile Data), Diversity(Alpha, Beta), Sample Clustering et cetera.