NGS(RNA-Seq)
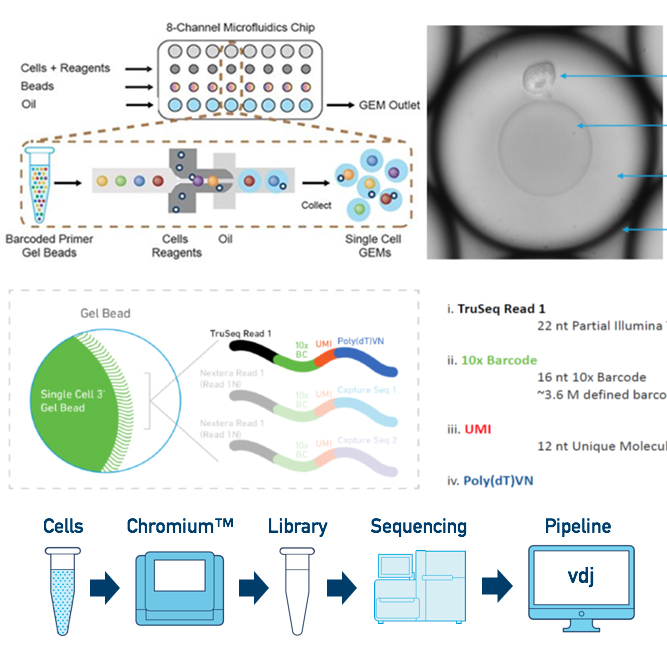
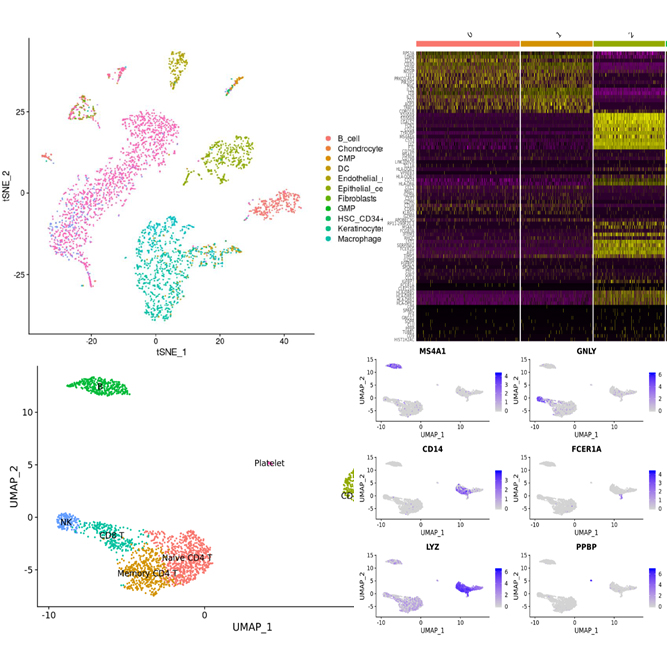
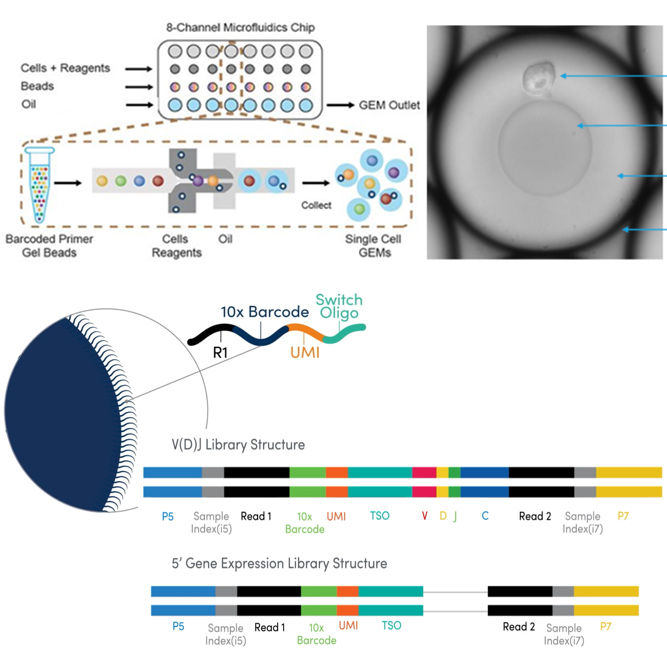
We offer single cell RNA-Seq service to analyze transcriptome of single cell for overcome the limit of whole transcriptome analysis of cell or tissue. Through this service, we can provide researchers with cell type identities, gene expression diversity of each cell and various data analysis such as UMAP, featrue selection, clustering et al..
Service Info.
| Sample requirement++ | - Cell line : ~1x106 cells in 14ml media(Cell viability >90%) - Primary cell or FACS sorted cell : 250,000 cells in 500 ul media (Viability >70%) |
|---|---|
| Library method | 10X Genomics Next GEM technology |
| NGS run format | Nova-Seq 6000, PE100bp |
| Data yield++ | ~60,000 reads(30,000 Paired Reads) per cell | Number of Target cell++ | ~5,000 cells |
| Turnaround time | ~4 weeks after Cell counting & QC |
| Sample type | Cell, Nuclei |
++ We can change start cell number, target cell number and sequencing depth after discussing with customer.
- Cell Count & QC
- Library construction
- NGS
- Data Analysis
- Report
Data Analysis
We support various data analysis such as gene expression data, UMAP, tSNE, feature selection, clustering heatmap, PCA, cluster biomarkers, cell type annotation, trajectory inference, cell-cell interaction, et al. with Cell Ranger, Loupe, Seurat, ExSCERA and other programs.
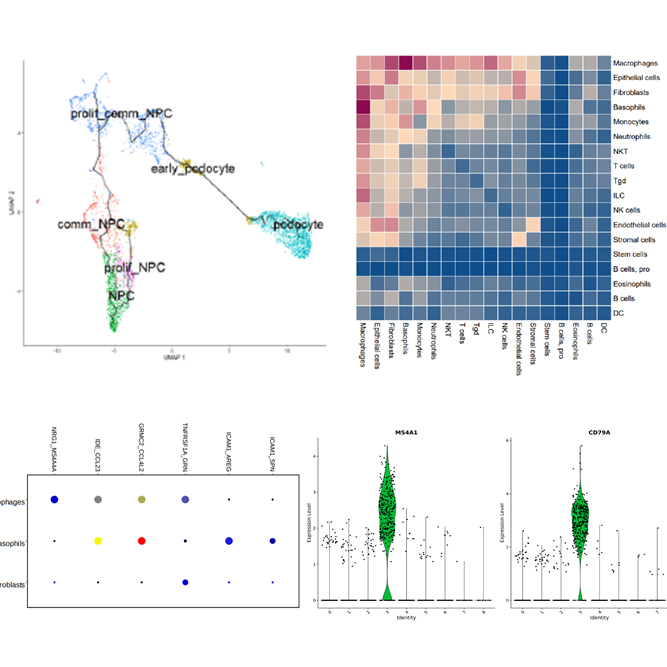
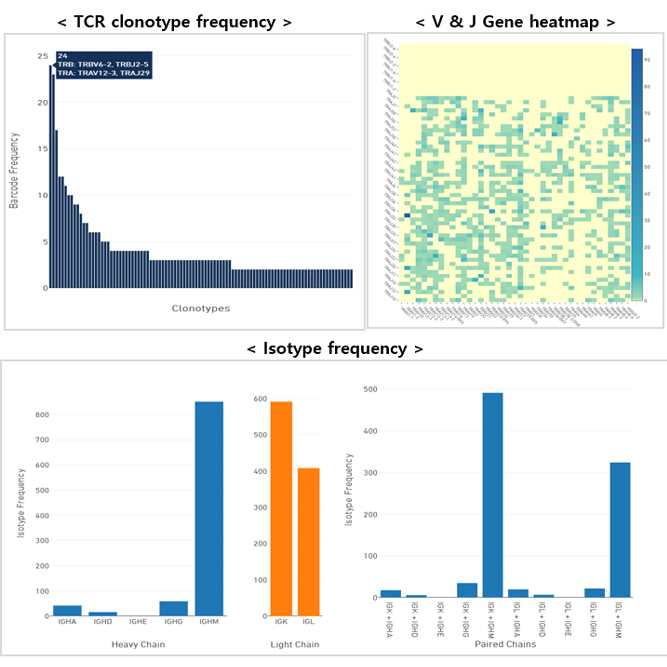
Single Cell Immune Profiling service provides a comprehensive approach to simultaneously examine cellular heterogeneity of the immune system, T- and B-cell repertoire diversity, and antigen specificity at single cell resolution. Through this service, we can provide researchers with cell type identities, gene expression diversity of each cell and various data analysis such as UMAP, Feature selection, Violin Plot. In addition, we support a variety of analyses such as clonotype determination by cell type, clonotype frequency for TCR and BCR, Full-length V(D)J gene sequences and heavy-light chain & α-β chain pairing.
Service Info.
| Sample requirement++ | - Cell line : ~1x106 cells in 1.5ml tube with media (Cell viability >90%) - Primary cell of FACS sorted cell : 250,000 cells in media (Cell viability >70%) |
|---|---|
| Library method | 10X Genomics Next GEM technology |
| NGS run format | Nova-Seq 6000, PE100bp |
| Data yield++ | * Minimum 15,000 reads pairs per cell for V(D)J library * Minimum 50,000 reads pairs per cell for 5’ Gene Expression library |
Number of Target cell++ | ~10,000 cells |
| Turnaround time | ~4 weeks after Cell counting & QC |
| Sample type | Cell |
++ We can change start cell number, target cell number and sequencing depth after discussing with customer.
- Cell Count & QC
- Library construction
- NGS
- Data Analysis
- Report
Data Analysis
We support various data analysis such as gene expression data, UMAP, t-SNE, feature selection, clustering heatmap, PCA, clustering heatmap, PCA, cluster biomarkers, cell type annotation, clonotype frequency for TCR and BCR, Full-length V(D)J gene sequences and heavy-light chain & α-β chain pairing.
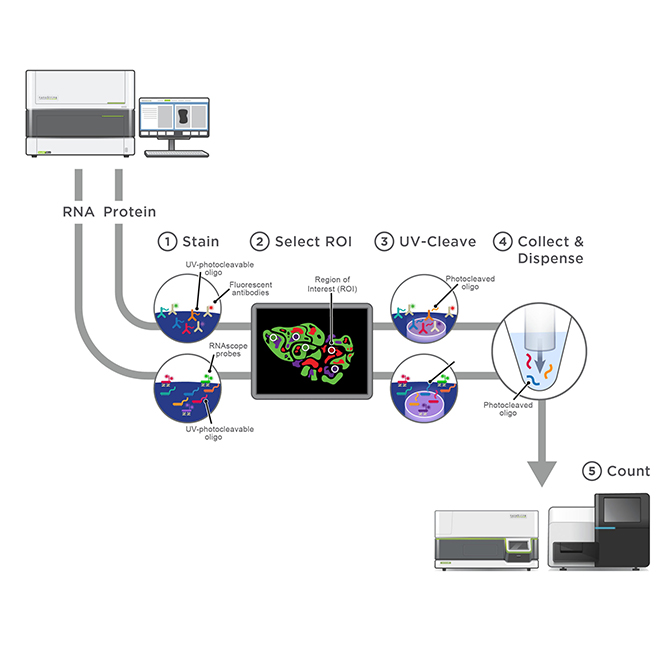
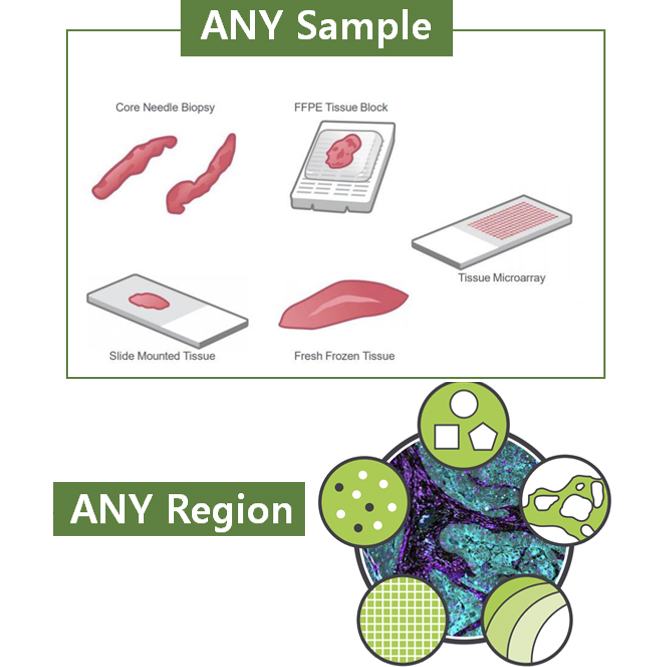
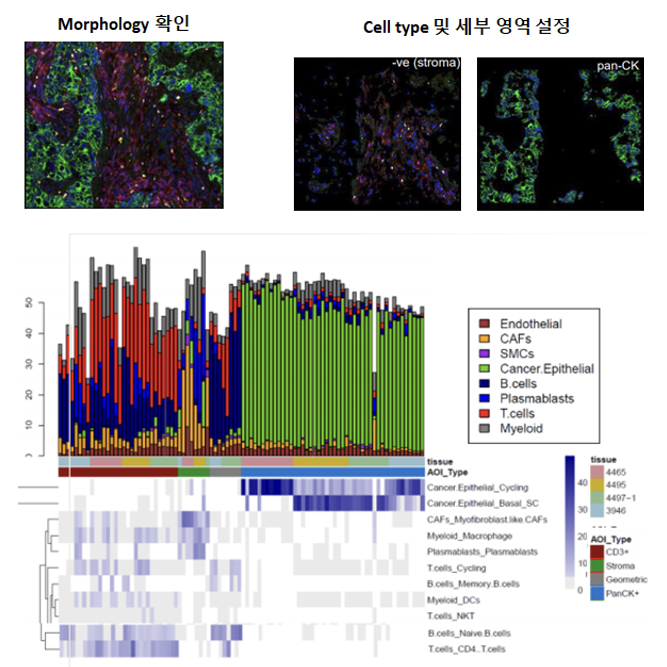
GeoMx Spatial Transcriptome Service combines the best of spatial and molecular profiling technologies by generating a whole tissue image at single cell resolution and digital profiling data. This unique combination of high-plex, high-throughput spatial profiling enables researchers to rapidly and quantitatively assess the biological implications of the heterogeneity within tissue samples.
Service Info.
| Service Name | GeoMx Spatial WTA* Service | GeoMx Spatial CTA* Service |
|---|---|---|
| Species | Human, Mouse | Human |
| Number of Gene | Human ~18,000ea / Mouse ~20,000ea | ~1,800ea |
| Data yield | ~100 reads/㎛2 | ~30 reads/㎛2 |
| Sample type | Tissue (FFPE, Tissue microarray, Fresh frozen tissue, Core needle biopsy etc,.) | Sample size | 14.6 x 36.2 mm |
| Turnaround time | ~3 weeks after ROI selection | |
| Publication | NanoString GeoMx Publication | |
*WTA (Whole Transcriptome Atlas) / *CTA (Cancer Transcriptome Atlas)
- Service Consulting
- Staining
- ROI Selection
- Library Construction & NGS
- Report
Data Analysis
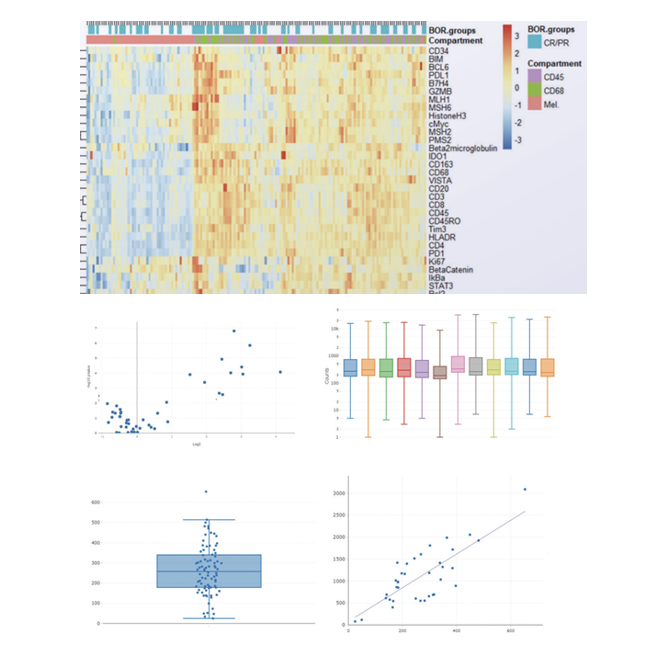
We support various data analysis such as connects Gene expression data(ExDEGA report) with spatial information. Also we provide visualization include clusters, heatmaps, volcano plots, bar graphs, box plots and scatter plots.
We offer transcriptome analysis by extracting RNA from biological species done with Genome Sequencing and applying NGS technology. In addition to mRNA expression profiling analysis, we support various analyses which include lncRNA, isoform, INDEL, Alternative Splicing Event and Gene fusion.
Service Info.
| total RNA-Seq | mRNA-Seq | |
|---|---|---|
| Sample requirement | >2ug total RNA | >2ug total RNA |
| Library method | RiboCop rRNA Depletion kit + CORALL RNA-seq Library Prep kit |
Poly(A) RNA Selection kit + CORALL RNA-seq Library Prep kit |
| NGS run format | HiSeq or NovaSeq, PE100 | HiSeq or NovaSeq, PE100 |
| Data yield | >6Gb/sample | >4Gb/sample |
| Turnaround time | ~4 weeks after RNA QC | ~4 weeks after RNA QC |
| Sample type | Tissue, Cell, Whole Blood(Paxgene), Plant, Fungi, Bacteria, etc. | |
++ If you want to get RNA prep.service, you can send us your samples after discussing with us.
- RNA QC
- Library prep.
- NGS
- Data Analysis
- Report
Data Analysis
| Basic Analysis | Optional Analysis | |||||
|---|---|---|---|---|---|---|
| Lnc RNA | mRNA Expression | Isoform Expression | INDEL/SNP Analysis | Splicing Variant | Gene Fusion | |
| Total RNA-Seq |
O | O | O | O | O | O |
| mRNA-Seq | O | O | O | O | O | |
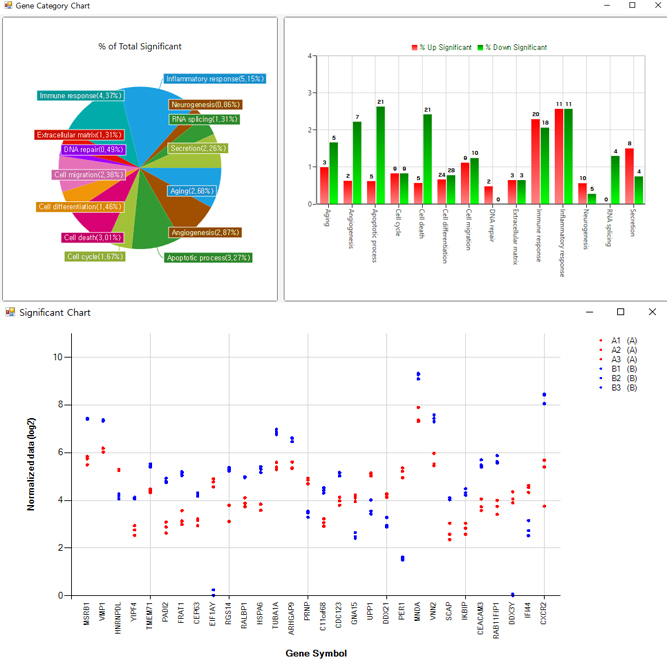
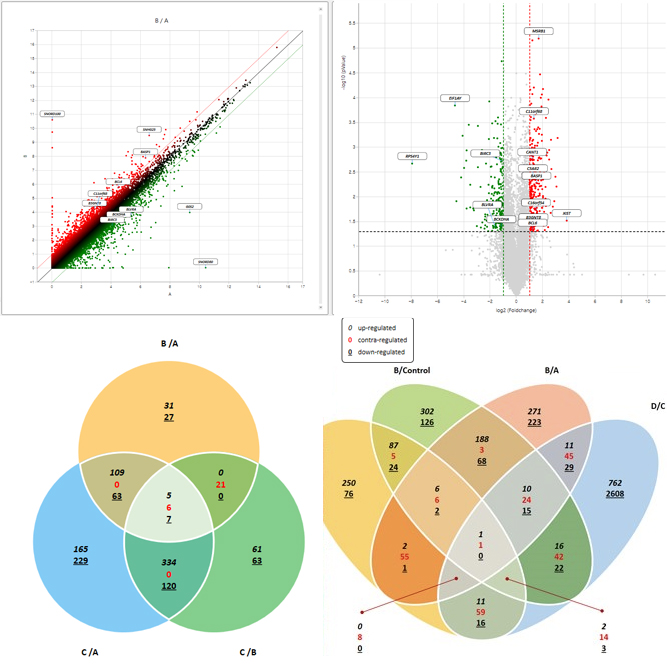
We support various analysis such as ExDEGA Report, GO, Pathway, Clustering, Network, GSEA, Data Mining et cetera..
We provide service of performing sequencing of 3’ UTR of RNA extracted from biological species with Poly(A) tail and analyzing gene expression. We provide high quality data with a low cost when only mRNA expression profiling analysis is requested.
Service Info.
| Sample requirement | >10ng total RNA |
|---|---|
| Library method | QuantSeq 3' mRNA-Seq Library Prep Kit FWD |
| NGS run format | NextSeq 500/550, SE 75bp |
| Data yield | >10M read/sample |
| Turnaround time | ~3 weeks after RNA QC |
| Sample type | Tissue, Cell, Whole Blood(Paxgene), Plant, Fungi, FFPE, Sorted cell, etc. |
++ If you want to get RNA prep.service, you can send us your samples after discussing with us.
- RNA QC
- Library prep.
- NGS
- Data Analysis
- Report
Data Analysis
We support various analysis such as ExDEGA Report (mRNA expression), GO, Pathway, Clustering, Network, GSEA, Data Mining et cetera..
Bacteria RNA-Seq is an important factor in data quality by efficiently removing only rRNA from total RNA. We can remove bacterial rRNA effectively using single-stranded DNA probes and provide customer with reliable transciptome analysis through NGS technology.
Service Info.
| Sample requirement | >2ug total RNA |
|---|---|
| Library method | RiboCop rRNA Depletion kit + CORALL RNA-seq Library Prep kit |
| NGS run format | HiSeq or NovaSeq, PE100(Paired End 100bp) |
| Data yield | >~2Gb (Data yield is varied depending on the species) |
| Turnaround time | ~4 weeks after RNA QC |
| Available species | Genome sequencing completed prokaryotes on NCBI DB (Species Search) |
++ If you want to get RNA prep.service, you can send us your samples after discussing with us.
- RNA QC
- Library prep.
- NGS
- Data Analysis
- Report
Data Analysis
We support various analysis such as ExDEGA Report (mRNA expression), GO, Pathway, Clustering, Network, GSEA, Data Mining et cetera..
We analyze, through NGS Sequencing, the amount of small-sized RNA such as microRNA and piRNA that exist in cell, tissue, exosome, and body fluid. Based on such database, we perform analysis of target gene and various kinds of data.
Service Info.
| Sample requirement | >2ug total RNA (Tissue, Cell) >20ng RNA (Exosome, Bodyfluid) |
|---|---|
| Library method | NEBNext Multiplex Small RNA Library Prep Kit |
| NGS run format | NextSeq 500/550, SE 75bp |
| Data yield | >20M read/sample |
| Turnaround time | ~4 weeks after RNA QC |
| Sample type | Tissue, Cell, Whole Blood(Paxgene), Serum, Plasma, Urine, Exosome, etc. |
++ If you want to get RNA prep.service, you can send us your samples after discussing with us.
- RNA QC
- Library prep.
- NGS
- Data Analysis
- Report
Data Analysis
We support various analysis such as ExDEGA Report (miRNA expression), piRNA profiling, GO, Pathway, Clustering, Network, Target gene, Data Mining et cetera..